A team of researchers, led by scientists at the University of Rochester, has identified a location in a gene of the influenza A virus that could be used as a "switch" for disrupting replication of the virus. If a way can be found to manipulate the switch in an organism, the researchers believe it would have important implications for stopping the spread of influenza.
The work was conducted by Professor of Chemistry Douglas Turner and scientists Walter Moss, Lumbini Dela-Moss, and Salvatore Priore at the University of Rochester, and chemists Ryszard Kierzek and Elzbieta Kierzek at the Polish Academy of Sciences in Poznan. Their findings were recently published in the journal PLoS ONE, produced by the Public Library of Science.
The scientists singled out a messenger RNA (mRNA) in their research because it allows the production of two proteins needed for viral propagation. Production of the second protein requires the mRNA to undergo the process of splicing, in which two remote sites of the long molecule join together, while the intervening segment is discarded.
"We look at one of the splicing sites as a potential switch," said Turner. "If we can inactivate the switch to prevent the two sites from coming together, we can stop the virus from spreading."
The scientists analyzed data from the sequencing of the influenza genome, which is available on a government database, in order to construct a 2-D rendering of the RNA molecule. By using a combination of computational and biochemical tools, they found that one of the splicing sites-referred to as 3'-exists in either an exposed hairpin loop or a difficult-to-access pseudoknot. The scientists concluded that there are two ways to prevent splicing of the RNA strands. One possibility is to hide the splicing sites from each other, which could involve forcing the 3' site to exist in the pseudoknot conformation. The second option would entail inserting a molecule into the 3' site to render it incapable of splicing. Either option, Turner said, would prevent production of a necessary protein and propagation of the virus.
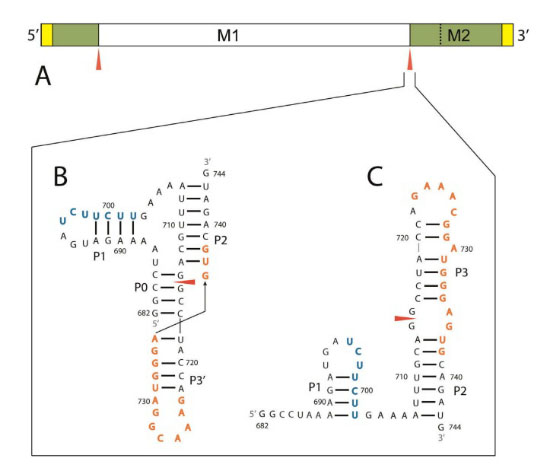
The 5' and 3' splice sites (red arrows) are shown in a basic rendering of a segment of the RNA molecule (A), as well as in the pseudoknot (B) and hairpin (C) conformations.
David Topham, co-director of the New York Influenza Center for Excellence, called the identification of a splicing switch an "unexpected discovery." Topham, who studies human immune responses to influenza infection and vaccination, said, "One can hope this will lead to novel antiviral drugs to treat severe influenza infection. This RNA-based research is only at the very beginning, but I see this as very important to the influenza field."
The researchers were originally taking a different approach to the influenza virus, which involved studying the role of RNA in viral packaging-the process by which a virus transmits its genome to a host cell.
"That proved to be a very difficult problem, so we changed our focus to the RNA structures in each segment of the virus," said Moss. "We found the switch from our calculations, and, fortunately, it had some interesting properties."
Turner's lab will now collaborate with Matthew Disney, associate professor of chemistry at the Scripps Research Institute, to explore the binding of known molecules to the influenza RNA. The hope is that the next phase of the research project will reveal important insights into the behavior of the virus, including how to stop the RNA splicing process.
